Sonador AI
Sonador AI provides an end-to-end suite of tools and best practice guidance for Medical Imaging based machine learning. It includes a framework for implementing machine learning models on top of PyTorch or Tensorflow, tools for creating annotated datasets, tracking experiments and model versions, deploying to production, and integrating AI into clinical workflows.
Manage the Machine Learning Lifecycle
Sonador AI is a collection of integrated tools and best practices to manage the complete machine learning. It provides software libraries that can be used to work with imaging meta and pixel data, three-dimensional representations of image stacks, interface with popular deep learning libraries such as PyTorch/MONAI and TensorFlow for creating models, and integrate with systems to help with their deployment and monitoring such as MLflow.
Explore and Visualize
Due to its power and flexibility, Jupyter has become the de-facto standard for Data Science development. It allows for developers and researchers to prototype complex workflows, validate results, and document design decisions in a single document.
Through its integration with Jupyter, Sonador provides an ideal environment for rapid prototyping AI models and preparing 2D and 3D medical imaging data. Data can be retrieved from Orthanc using the Python client library, converted to NumPy arrays using PyDICOM, analyzed using SimpleITK, and visualized with ITK Widgets.
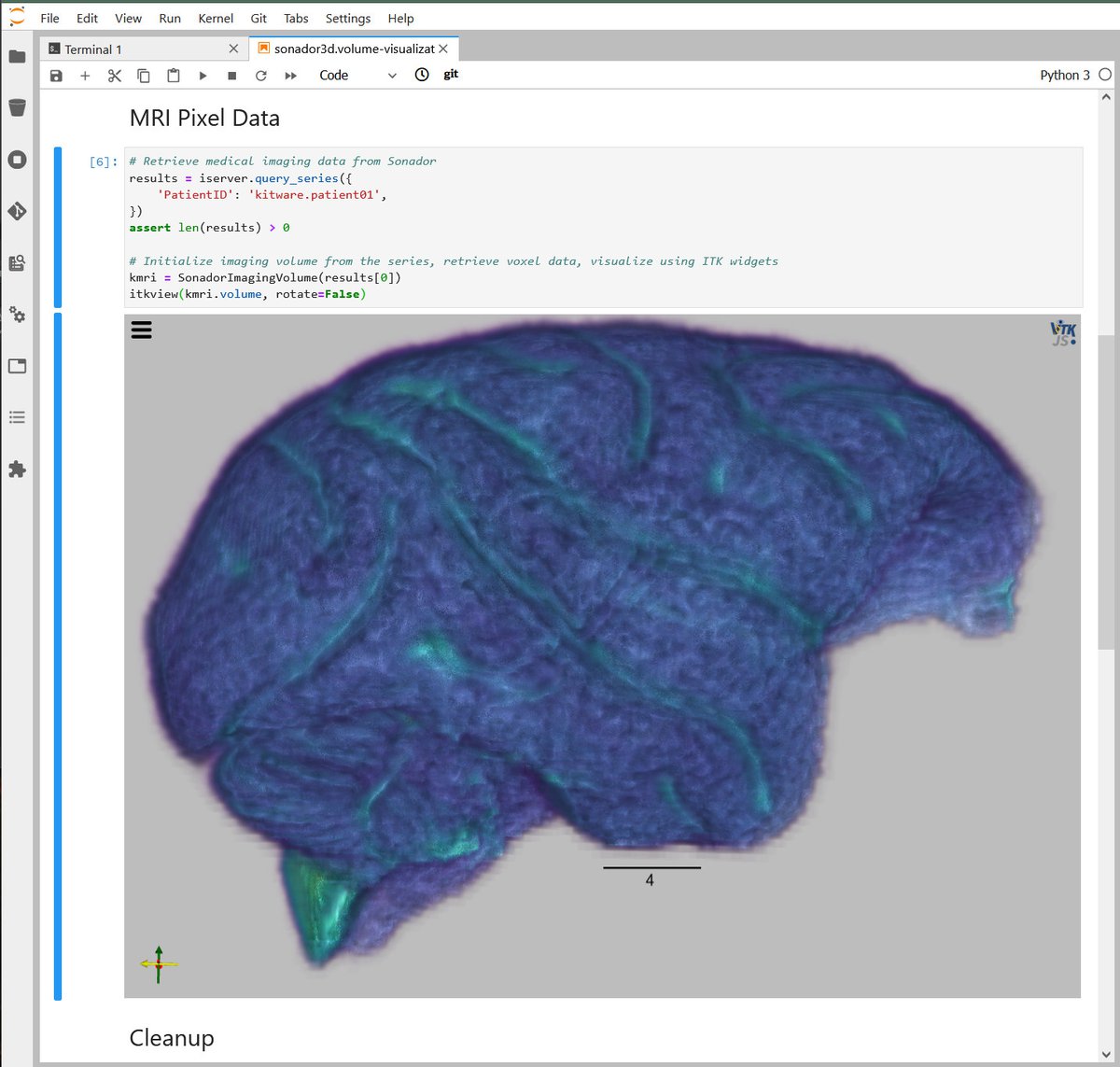
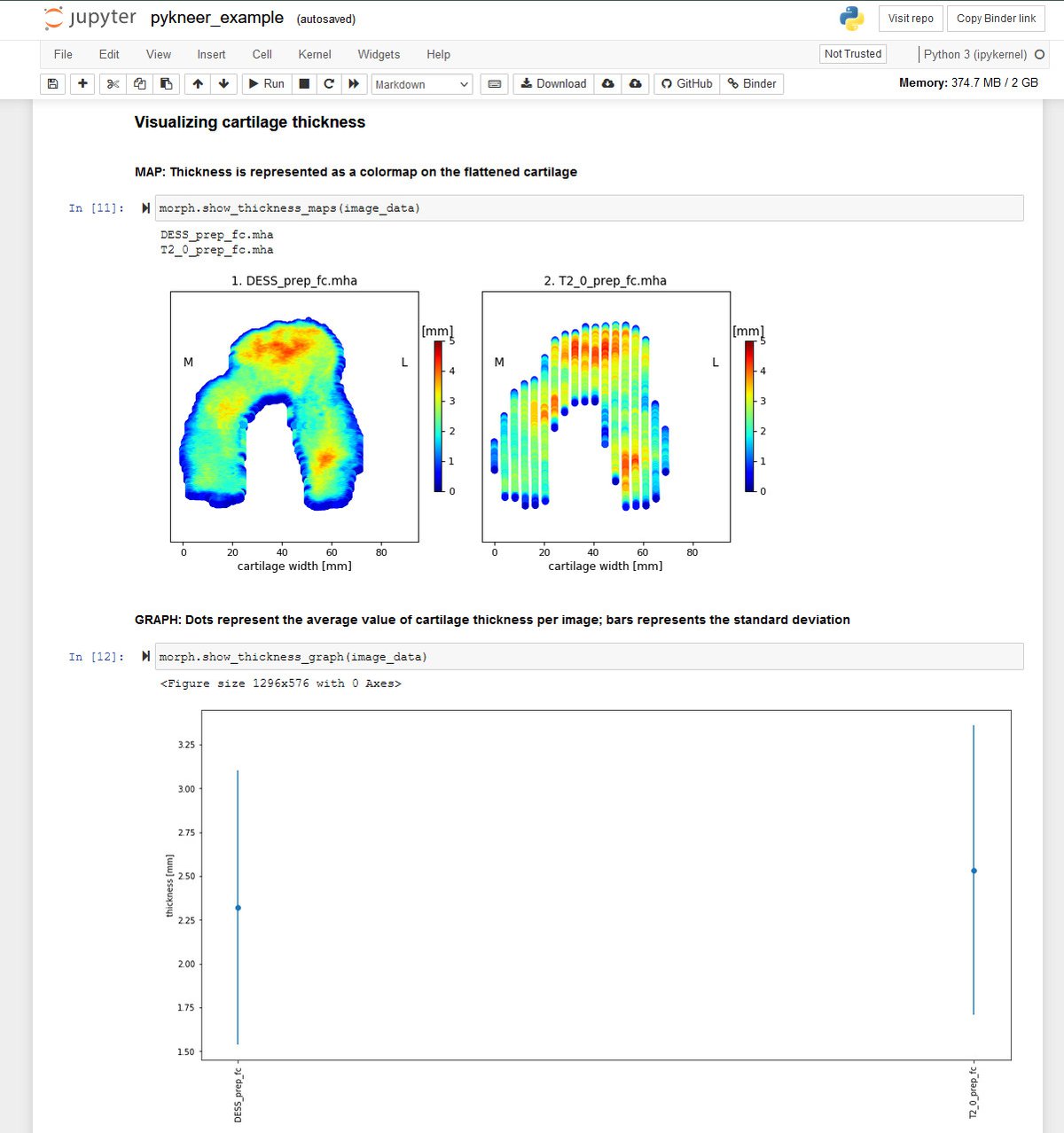
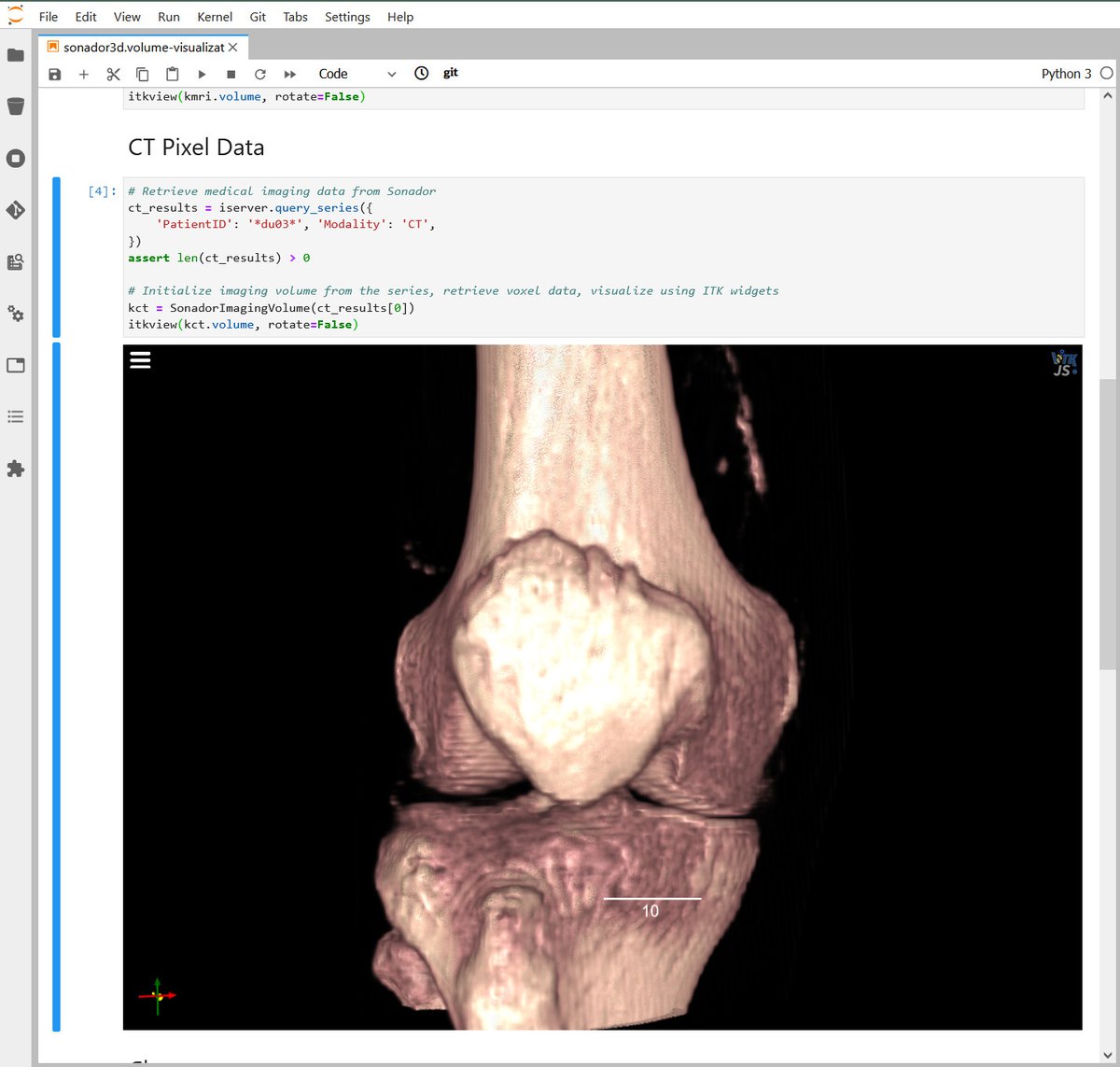
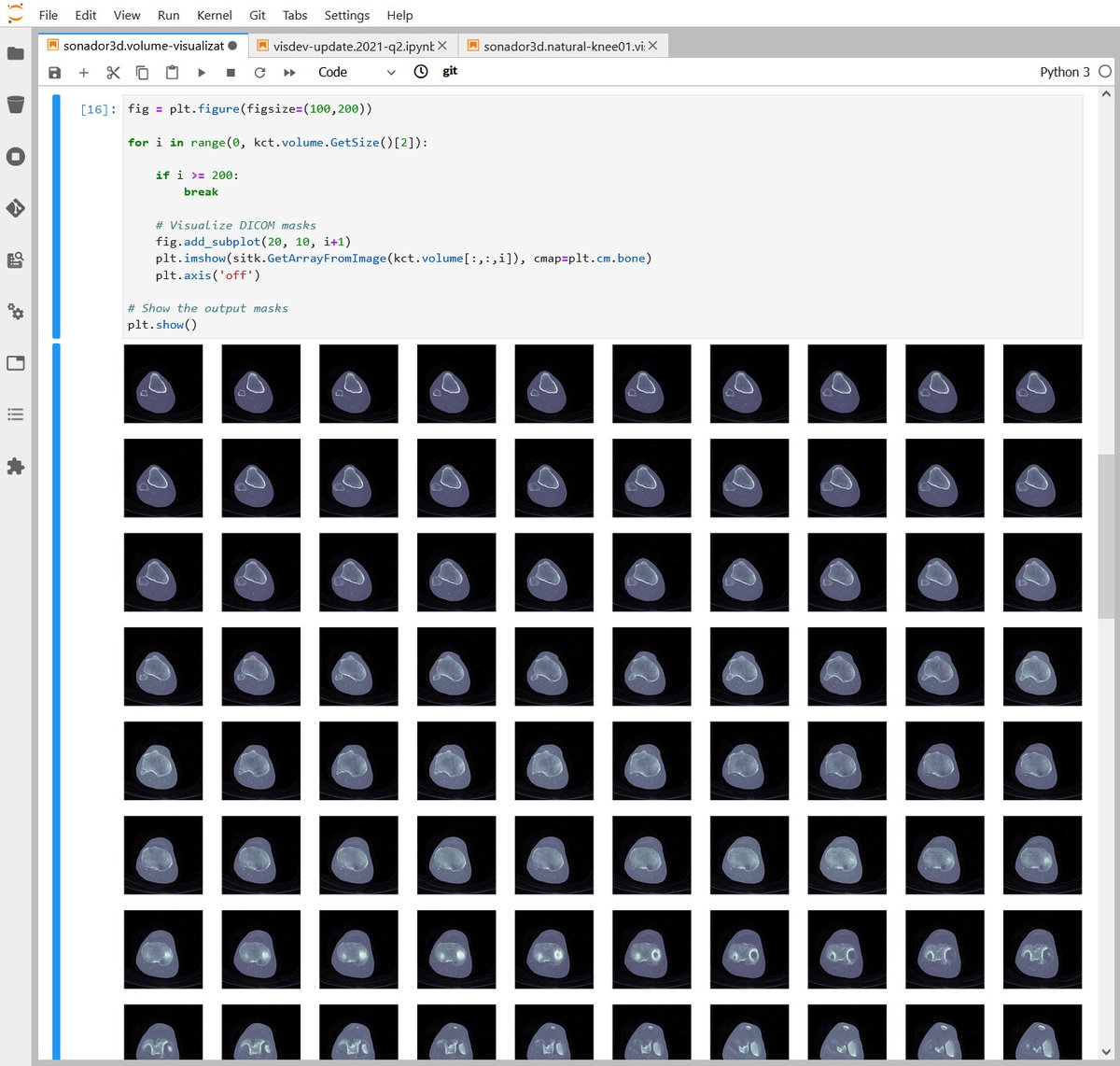
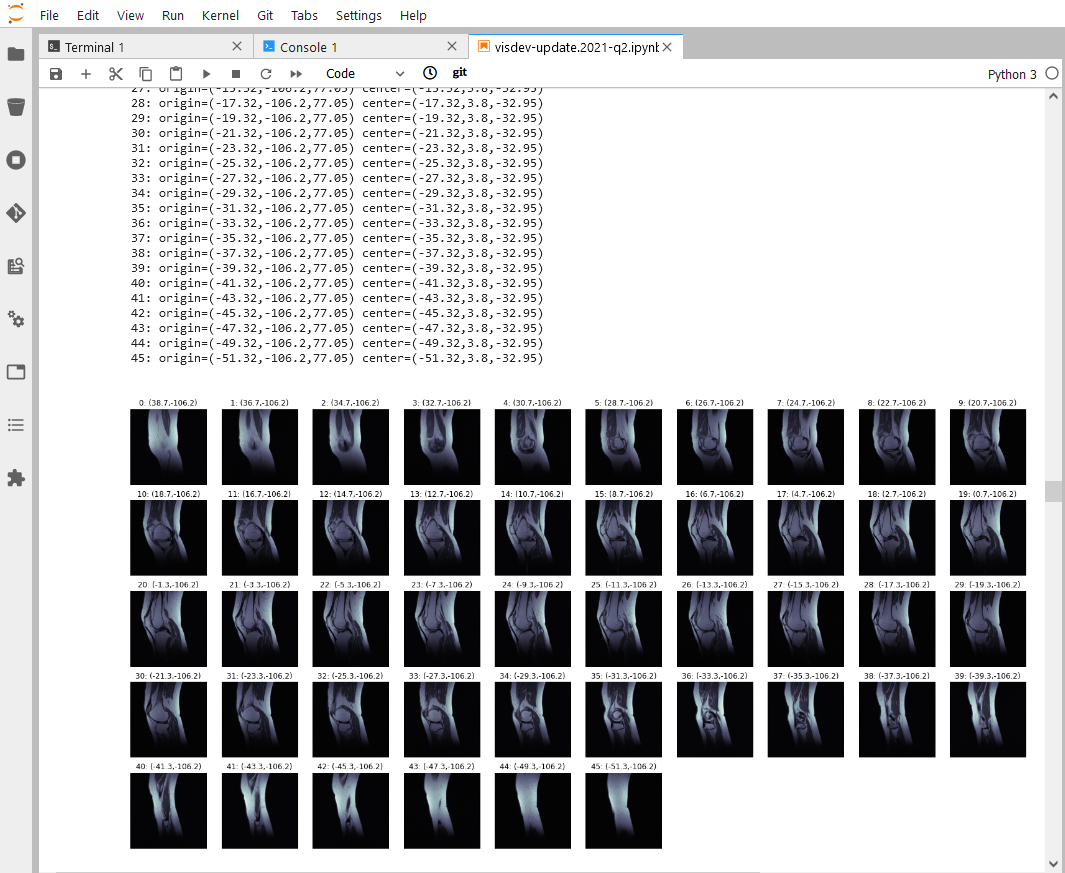
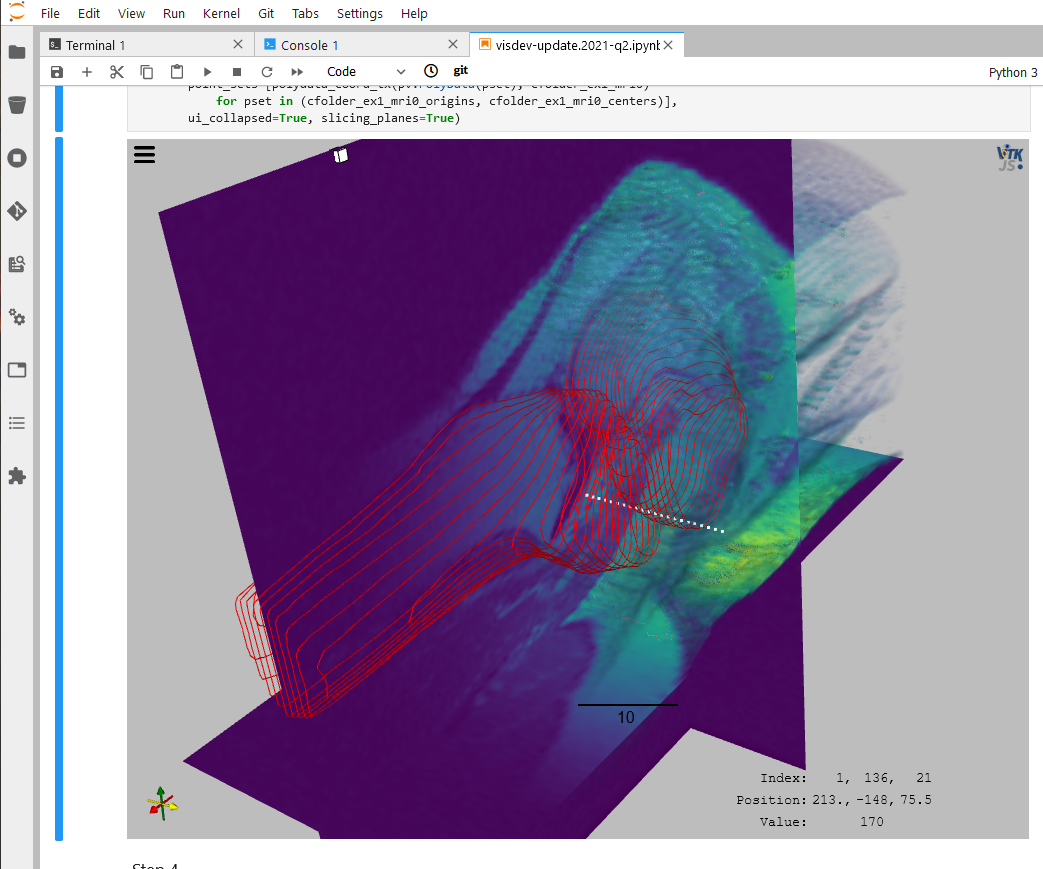
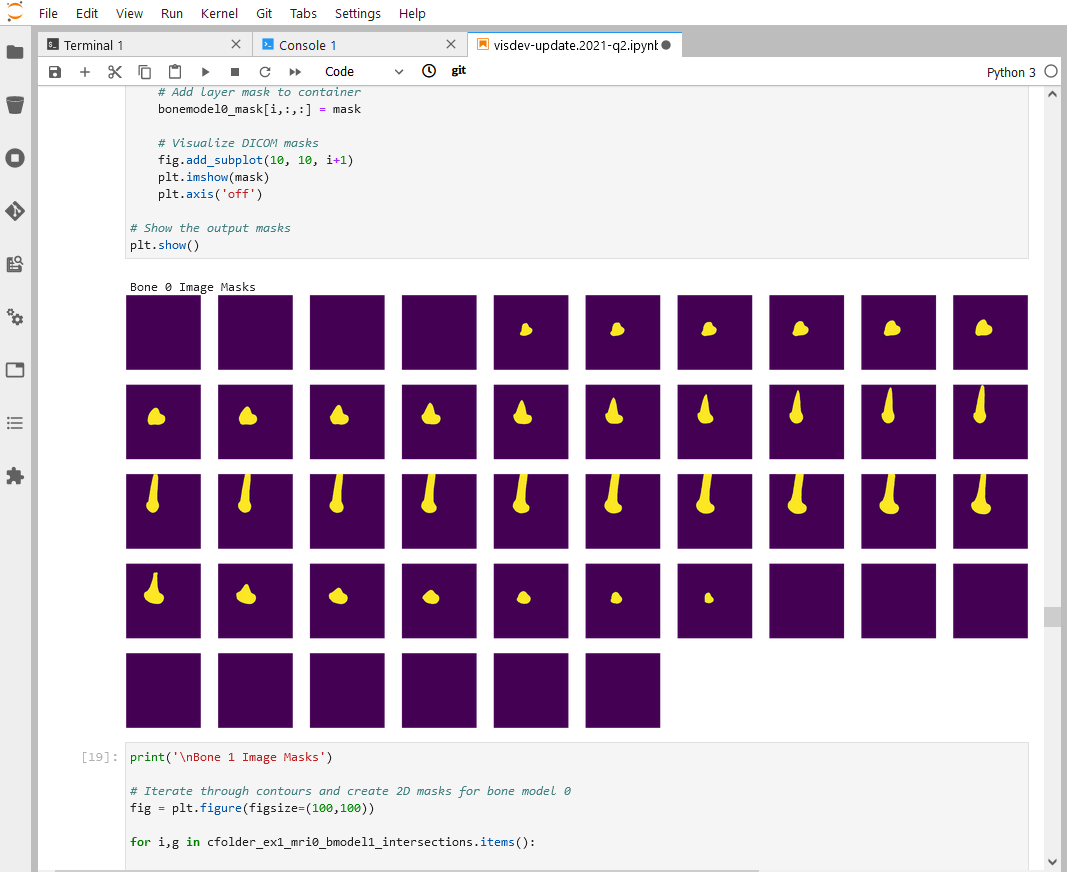
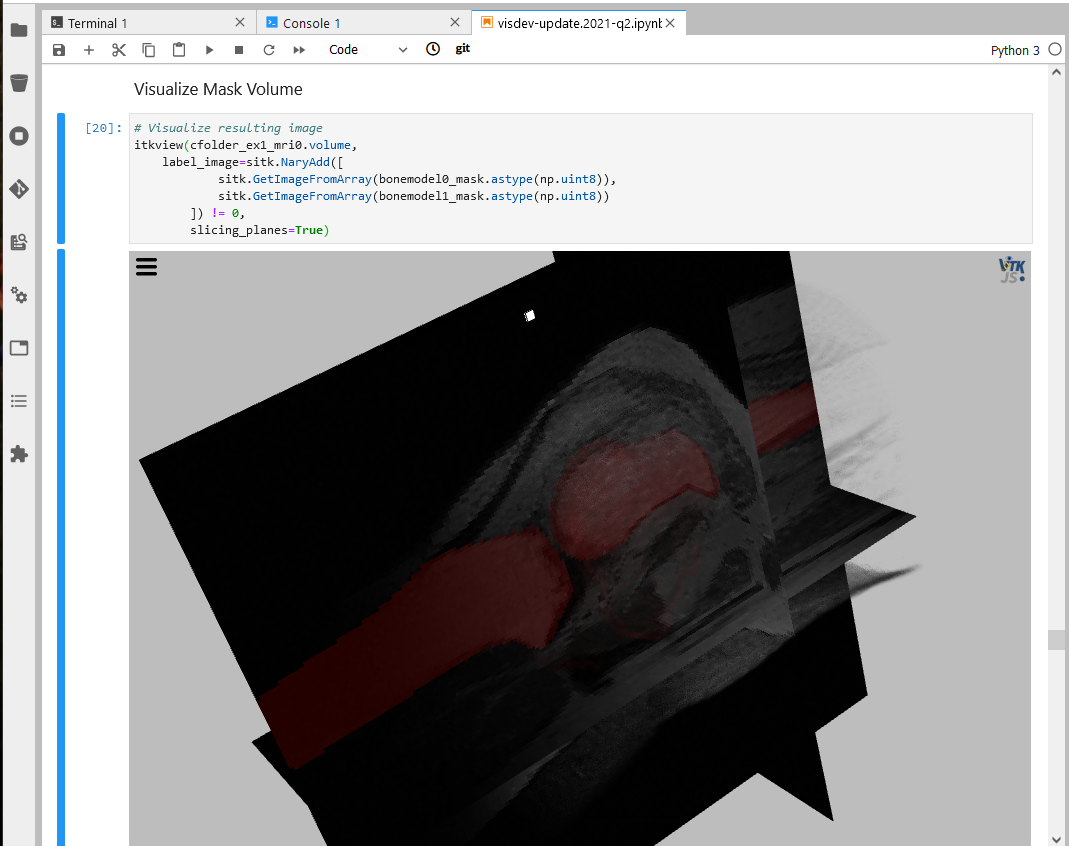

Create and Assess AI Models
Sonador is compatible with the Medical Open Network for Artificial Intelligence (MONAI) framework. MONAI is a collection of components and programs designed to build end-to-end AI systems based on medical imaging standards and best practices. Built on top of PyTorch, MONAI provides a framework for multi-dimensional medical imaging pipelines; implementations of networks, losses, and evaluation metrics; and support for multi-GPU training.
In addition to the core modules, MONAI also includes MONAI Label, a tool that enables researchers to build AI annotation models quickly. Available within 3D Slicer as a plugin, MONAI Label is able to observe as users segment or align structures of interest and then update a background model so that it becomes more accurate based on user feedback.
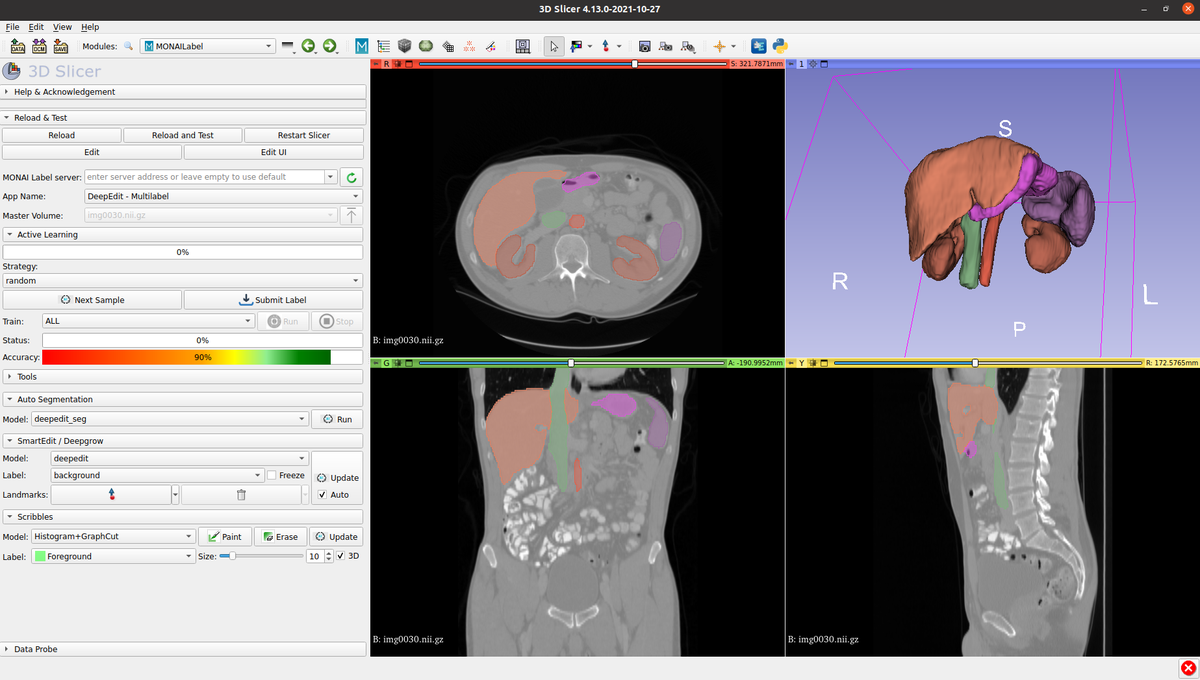
Verify and Validate
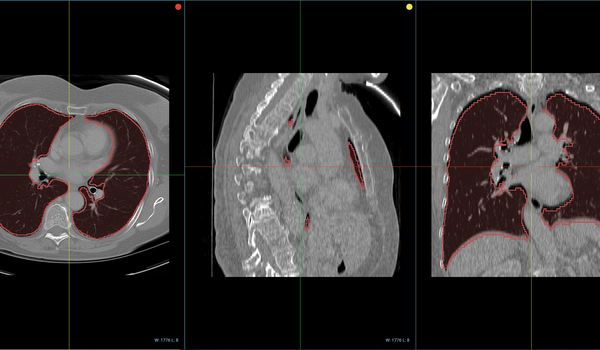
While a challenge in any industry, labeling Medical Imaging data for training and then verifying and validating model outputs is particularly difficult and time-consuming. As compared to other industries, where a layperson might label an image as "car" or "street" without too much trouble, reviewing data in medical imaging requires a professional's opinion.
To streamline labeling and review tasks, OHIF provides a powerful set of annotation tools to capture measurements, findings, or other features of interests. The annotations can be paired with the original study and saved as DICOM-SR documents and sent back to Orthanc, enabling them to be transferred along with the source images for downstream processing.
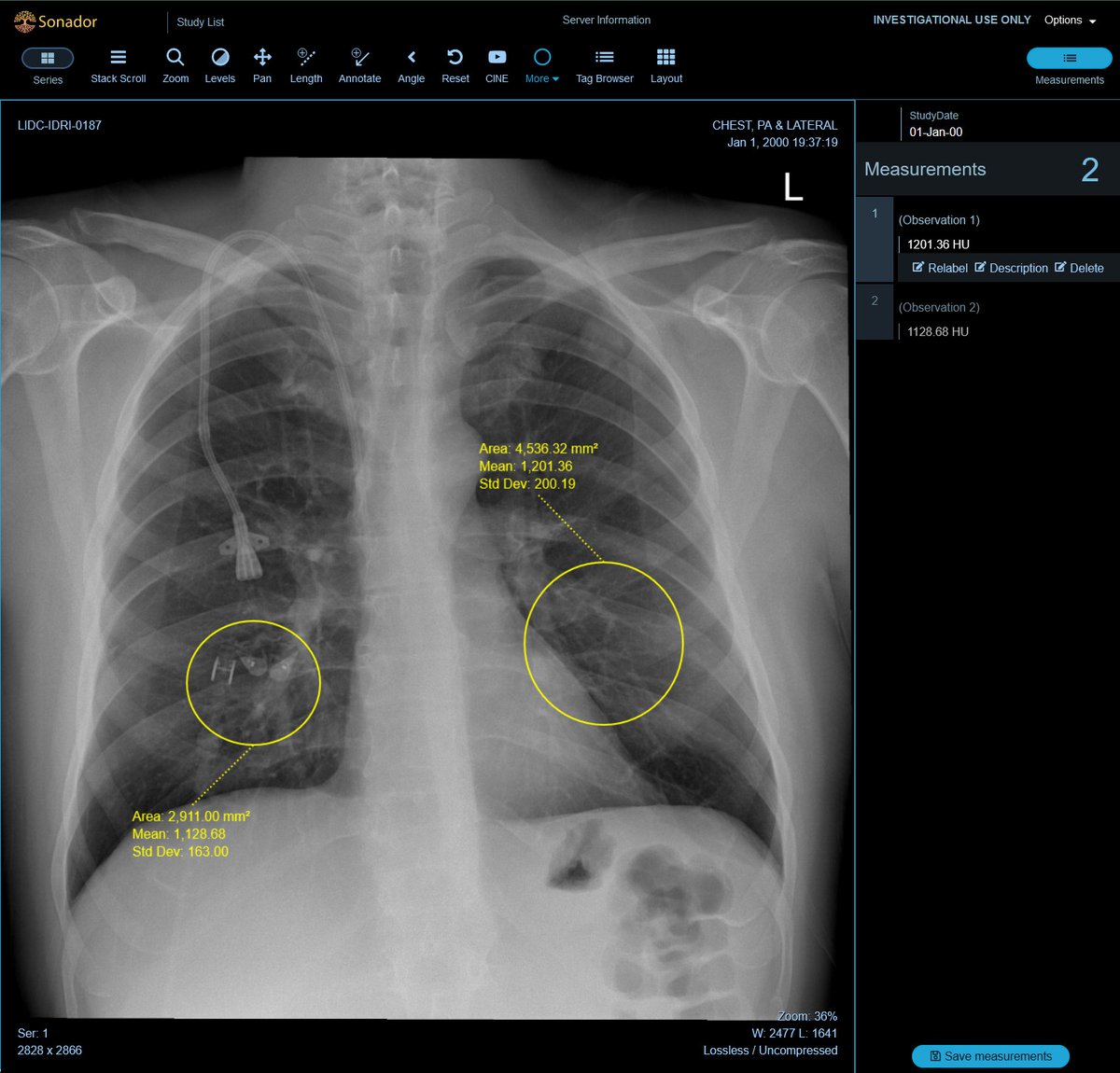
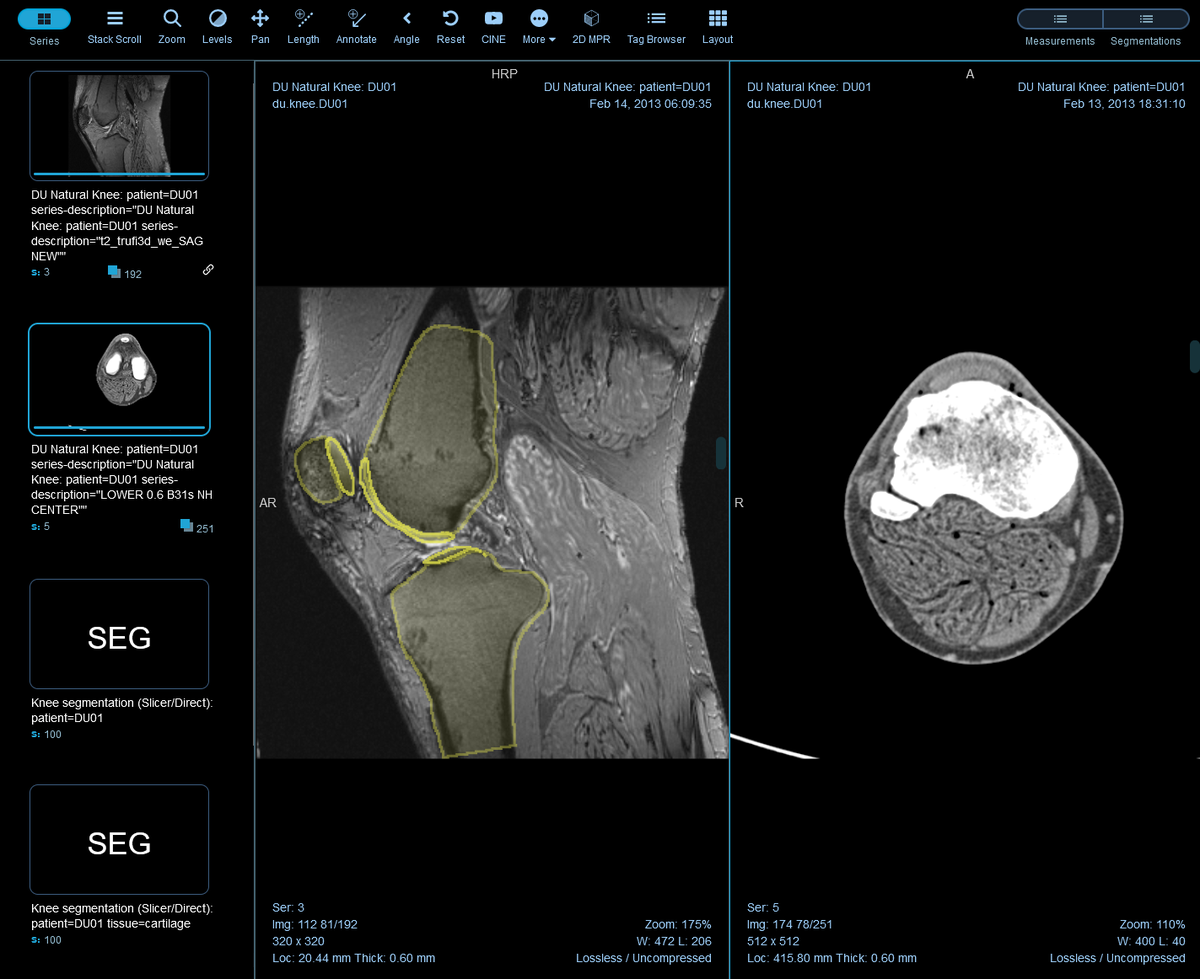
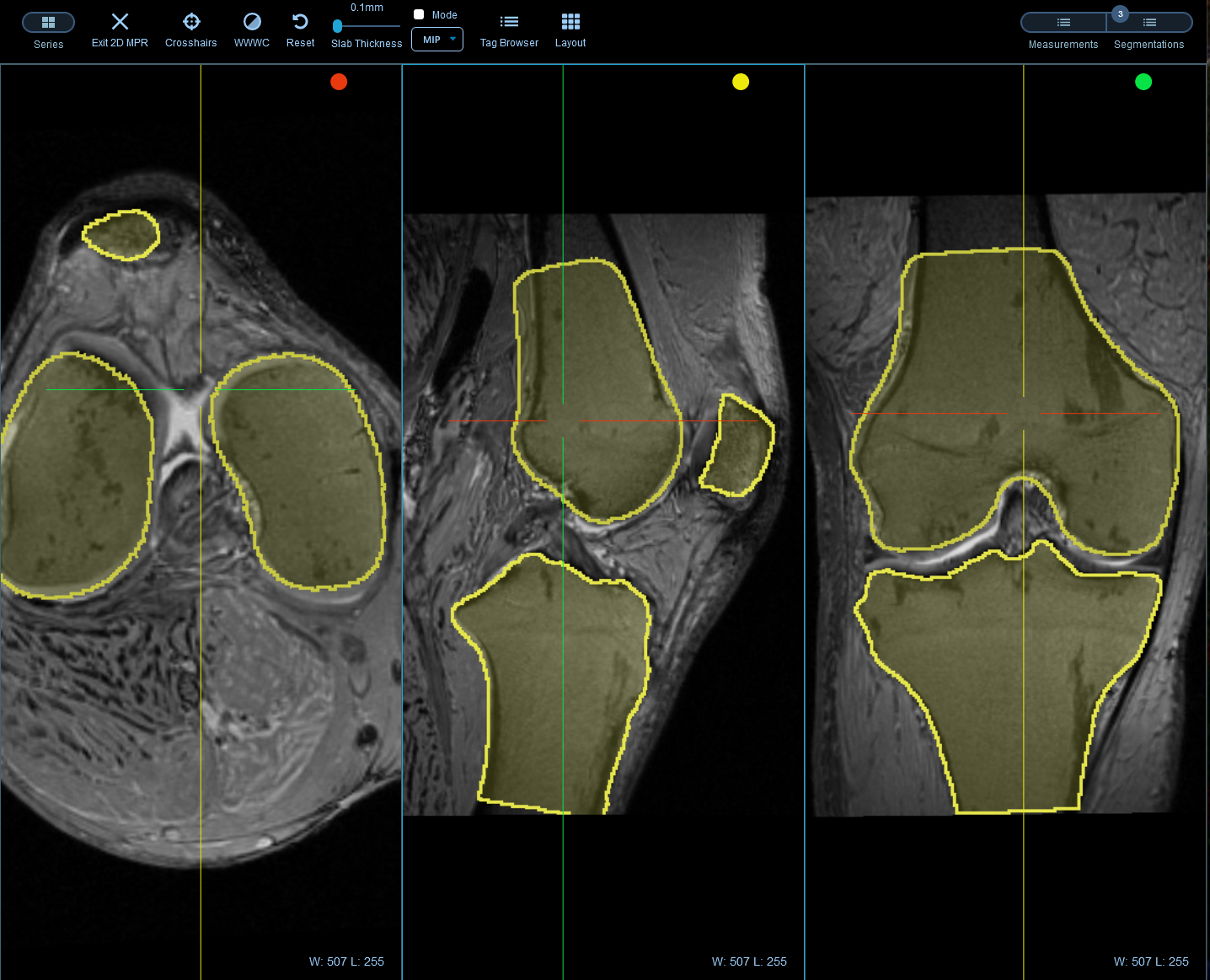

Publish and Deploy
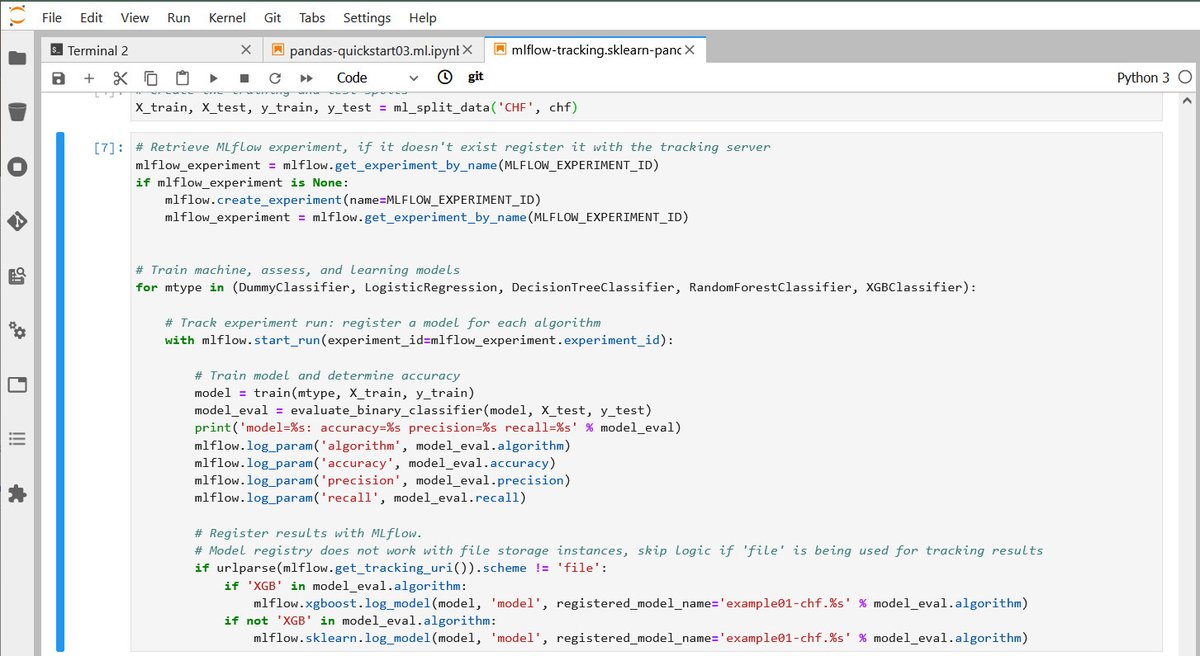
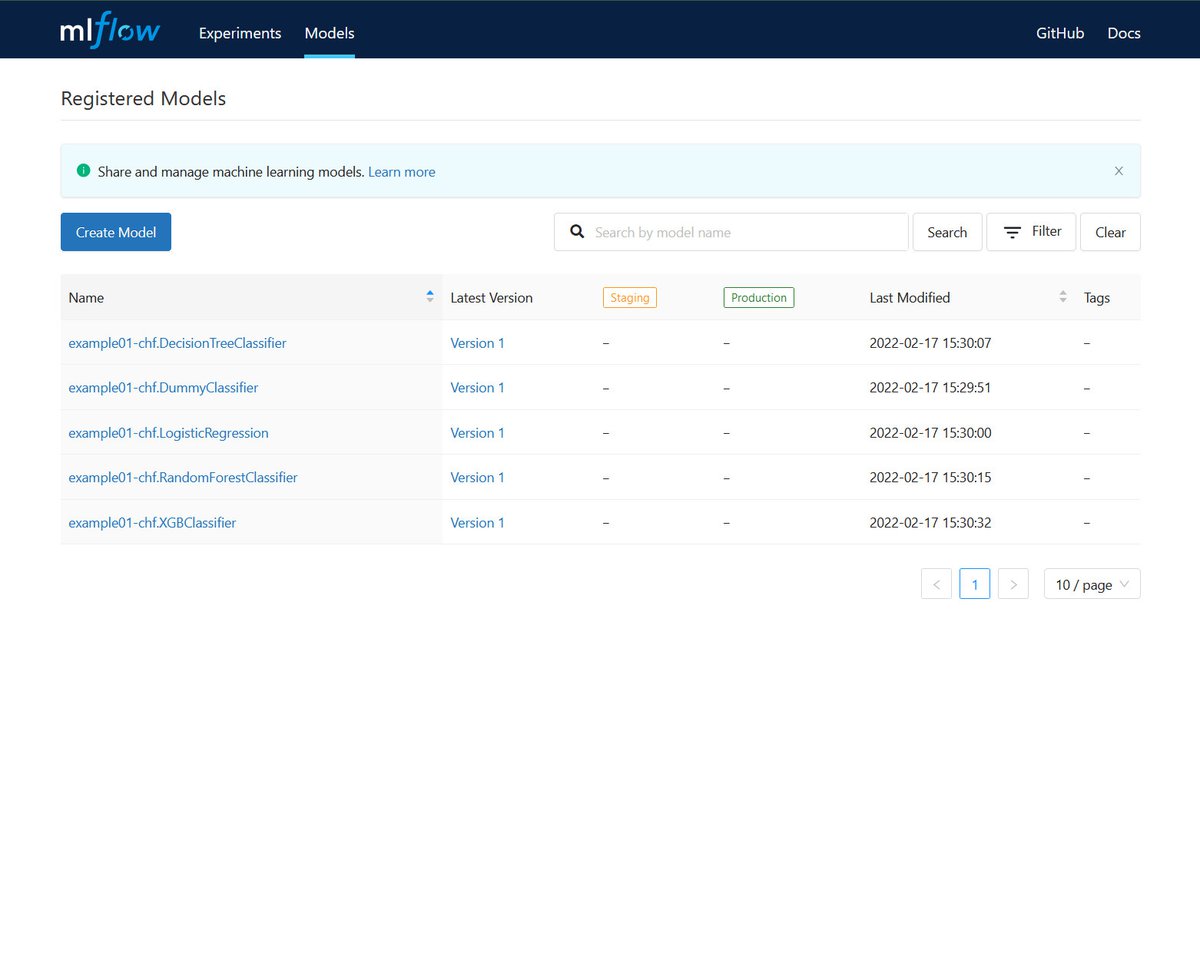
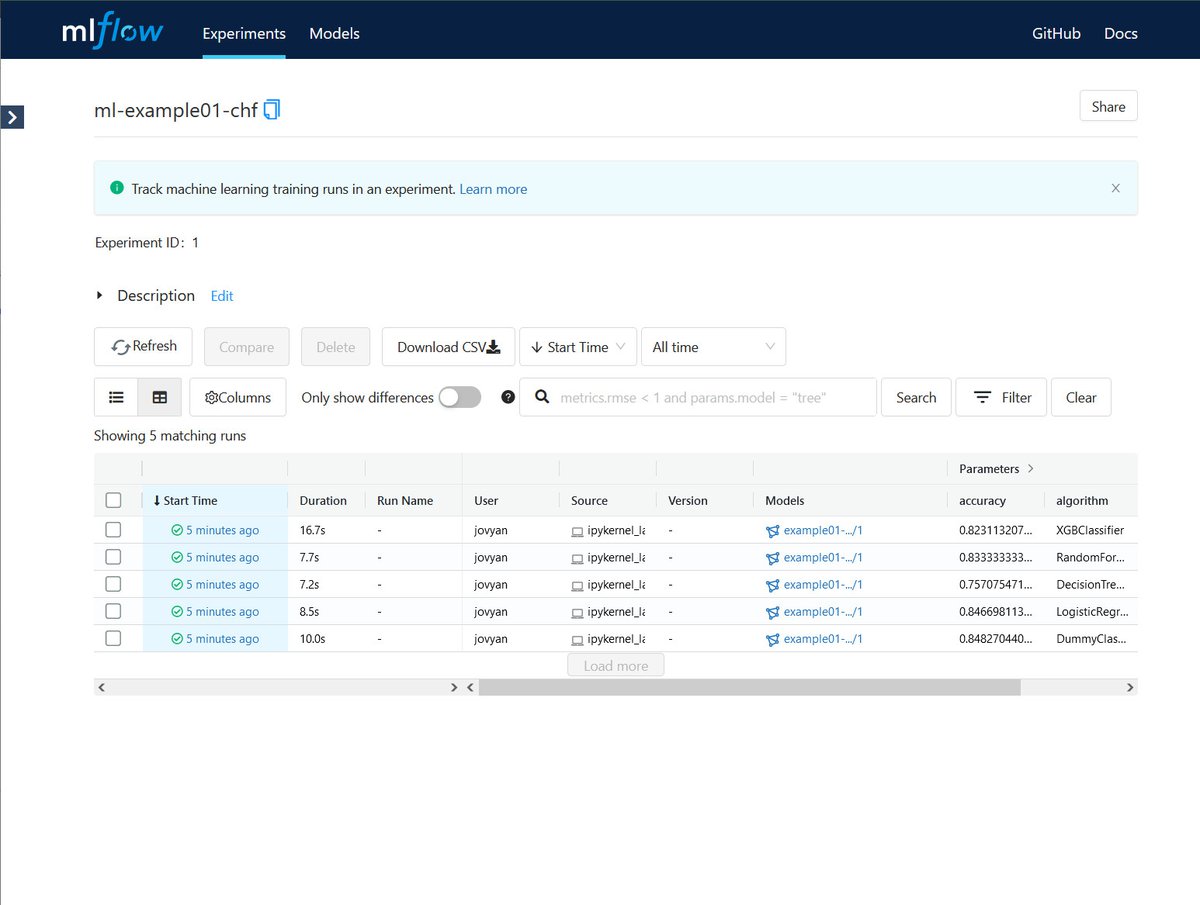
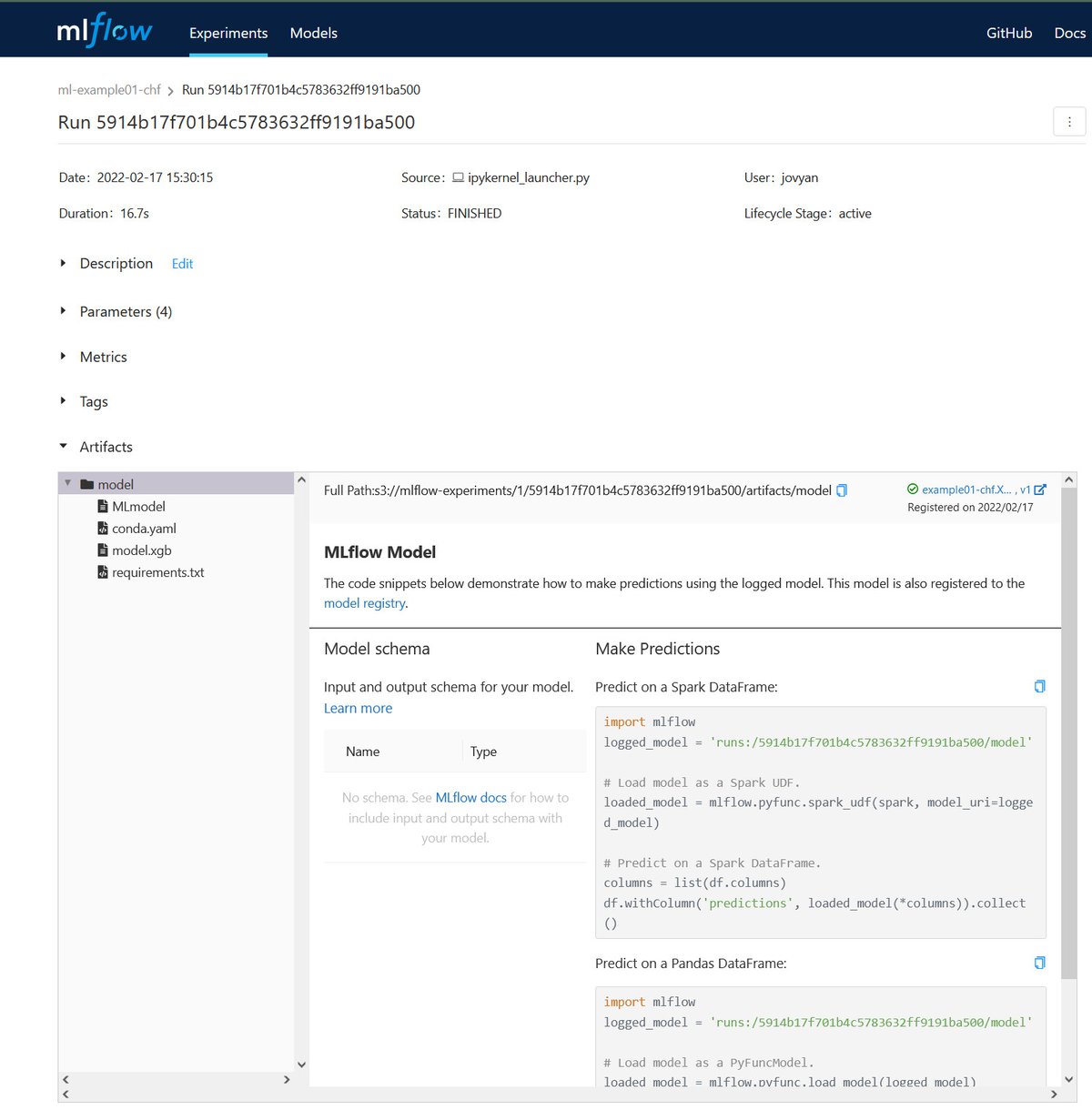
Sonador AI integrates with MLflow to provide tools to help manage the machine learning lifecycle. It provides a central system to track parameters during training to ensure reproducibility, host binary artifacts centrally to ease deployment, and track which models are deployed in what environments.
MLflow is designed to work with any machine learning library, algorithm, deployment tool, or language. It provides a set of REST APIs and simple data formats that allow for it to be integrated into a variety of tools including REST APIs, stream data consumers, or serverless functions.
Comments
Loading
No results found